Menu
Searching for building units and the underlying net in the crystal structures of ZrZn22 and NaCd2
Algorithm:
1. Convert the files ZrZn22.cif and NaCd2.cif into the ToposPro database Samson. Repeat the analysis for these records. (Set value Min. Omega=0.00 in Nanoclustering window)
2. In the case of ZrZn22 there are only three representations and each of them consists of two one-shell clusters; the two last representations contain “empty” cluster with the center in Wyckoff position 8b. Select the representations and check the option Generate Nets to create a new database that will contain the underlying nets.

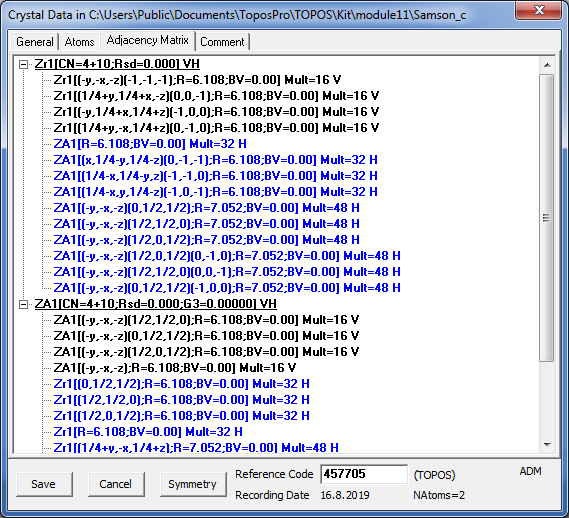
3. Open the Crystal Data window for the first record and click the Comment tab. You see the information on the cluster composition:

that means that atoms Zn1 and Zr1 in the underlying nets correspond to 13-atom and 17-atom clusters, respectively. Then go to the Adjacency Matrix tab and be sure that the cluster centered by Zn1 is in contact with 12 other clusters (6Zn1+6Zr1) while the other cluster is linked to 16 clusters (12Zn1+4Zr1).

Open the Crystal Data window for the second record to be sure that one of two clusters is “empty” (ZA1), moreover, some intercluster contacts in the adjacency matrix are marked as “H bonds”. Pay attention that in the underlying net each “valence” contact corresponds to a pair of “tangent” clusters having at least one common atom, while “H bond” contact designates “bounded” clusters that are connected only by bonds between their surface atoms.
4. Close the Crystal Data window and run ADS for the first record in the Classification mode. You will find that the underlying net has the topology of the cubic Laves phase MgCu2 (mgc-x). The topology of other two underlying nets can be analyzed in two ways: ignoring or not ignoring links between “bounded” clusters. For this purpose you can set the Bond type option for H bonds to None or to At., respectively.

Check that for the second underlying net the two possible topologies are diamond (dia) and b.c.c. (bcu-x), respectively, while for the last underlying net the topologies are unknown to ToposPro.
5. The beginning of the Samson.txt file looks like:

6. Turn to the database Samson and run IsoCryst for the ZrZn22 record. Draw the picture and select all Zr1 and Zn3 atoms.

Leave the centers of clusters only and press one time the Growth button to be sure that the whole structure can be constructed with one-shell clusters centered by Zr1 and Zn3 atoms. Select Zr1 atoms only and show their coordination polyhedra ( ) to see them connected by vertices. Do the same for the Zn3 coordination polyhedra.
) to see them connected by vertices. Do the same for the Zn3 coordination polyhedra.


7. Pay attention that the number of the NaCd2 nanocluster representations is much larger (41), but there is only one with two clusters (the simplest one). Select this representation only and construct its underlying net, be sure that the topology of the net is also mgc-x if both tangent and bounded cluster contacts are taken into account.

Open Samson.txt and be sure that both clusters are two-shell: Na1: 1@16@44 (61-atom cluster) and Cd6: 1@12@50 (63-atom cluster).
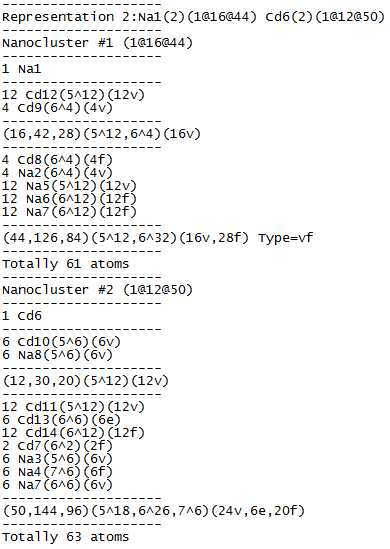
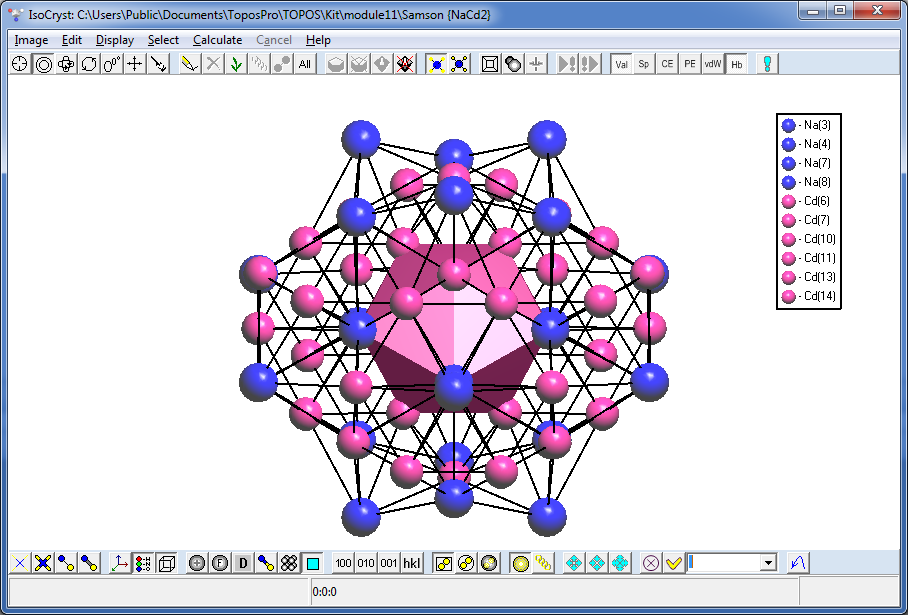
Draw both clusters in IsoCryst and look how the atoms of the second shell are allocated relative to the atoms of the inner polyhedron:

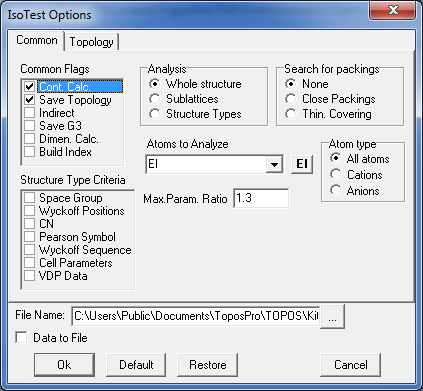
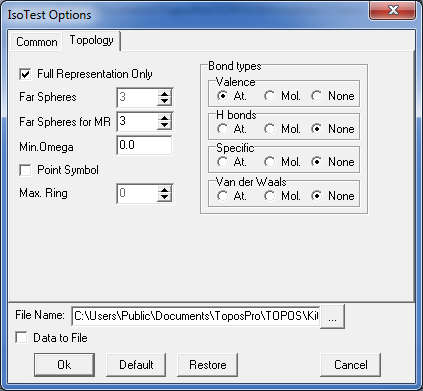
8. Select both records in the Samson database and open the IsoTest program. Click the option Common/Common Flags and indicate the Save Topology and Cont.Calc. mode. Then specify the following IsoTest options in Topology tab to compute coordination sequences for all atoms up to the 3rd coordination shell and store the information into the Samson.its file:
Run IsoTest and close its window after finishing the calculation. Then right-click in the database window and specify Show ITS to show information about coordination sequences in the Crystal Data window.

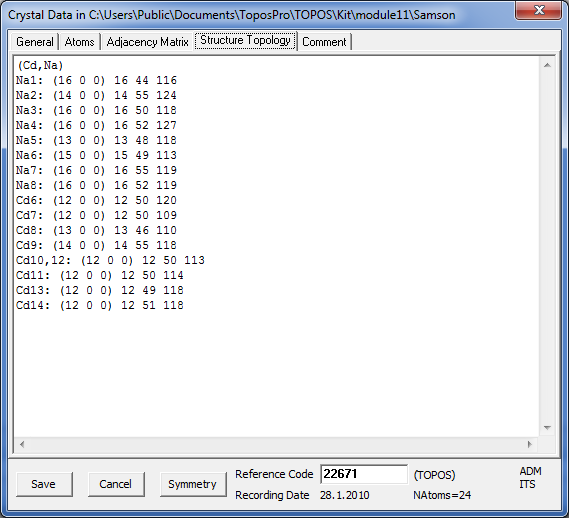
Open the Crystal Data window and go to the Structure Topology tab to see the coordination sequences.
Close the Crystal Data window and open the Distribution window (Database/Distribution). In the Distribution Type option set Sublatice Topology and press the Create Distribution button to construct the distribution of the coordination sequences.
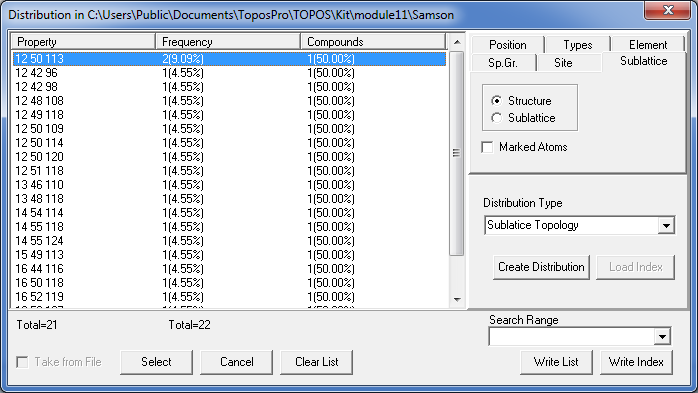
Order the list by Property.
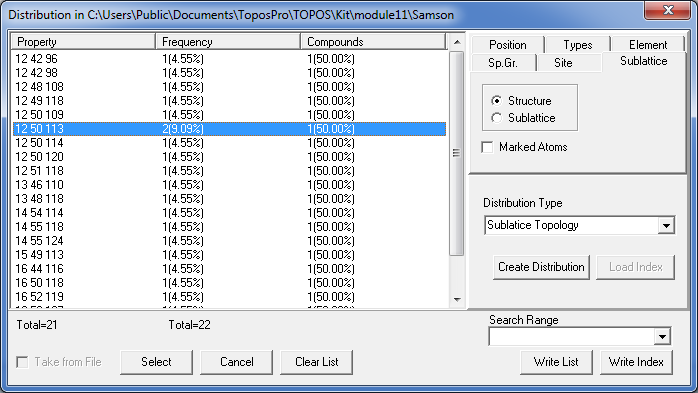
You see, for instance, that the clusters 1@12@50 allow four possible compositions for the third shell with 109, 113, 114 or 120 atoms, moreover, all of them occur in NaCd2 (you may press the Select button to select the corresponding records in the database).
9. Using the Crystal Data window, find that the cluster 1@12@50@113 is realized for Cd10 and Cd12 atoms of NaCd2. Open IsoCryst and construct three-shell cluster for Cd10 by two successive clicks of the Growth button. Select all atoms in the cluster and use Save Selection command (Graph files (*.gph) type of files) to store the cluster topology in the 1_12_50_113.gph file.

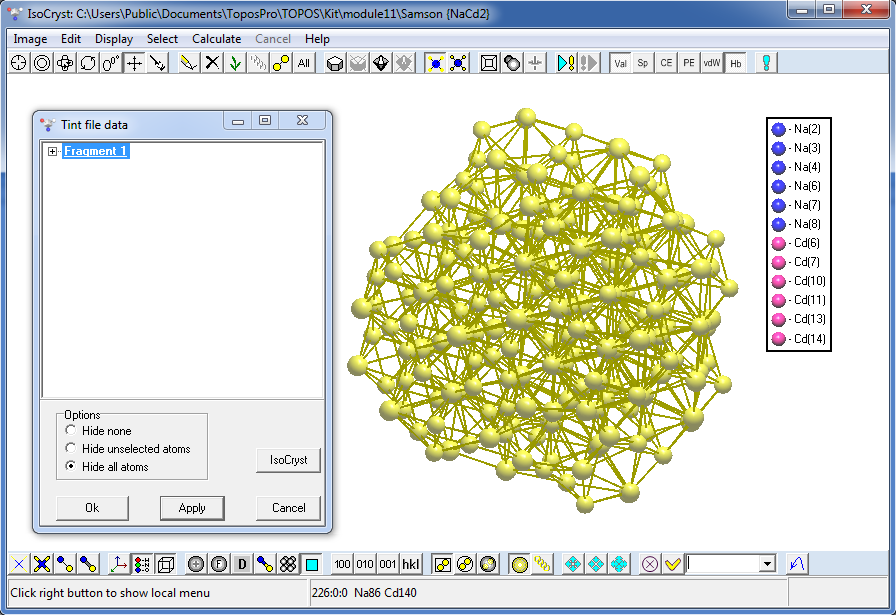
10. Open IsoCryst for the NaCd2 record, draw picture and use the Load Tint File command (Graph files (*.gph) type of files) to choose the 1_12_50_113.gph file to find that there is only one independent cluster of this type.

Press the Apply button to show visible fragment of the cluster, press the IsoCryst Growth button several times until the Fragment item in the list becomes bold (all atoms are visible).

Press the Apply button again and center the image (Space key) to see the cluster required.
Exercise: perform the same analysis for the crystal structure of β-Mg2Al3 (Mg2Al3.cif). What are the relations between this structure and the structures of ZrZn22 and NaCd2? Note: also use MinOm=0.00 value in Nanoclustering window.
(The nanocluster model can be presented as a combination of 1@16@44 and 1@12@50 nanoclusters with the Mg9 and Al14 centers, respectively. The topology of the net is mgc-x).

Algorithm:
1. Convert the files ZrZn22.cif and NaCd2.cif into the ToposPro database Samson. Repeat the analysis for these records. (Set value Min. Omega=0.00 in Nanoclustering window)
2. In the case of ZrZn22 there are only three representations and each of them consists of two one-shell clusters; the two last representations contain “empty” cluster with the center in Wyckoff position 8b. Select the representations and check the option Generate Nets to create a new database that will contain the underlying nets.

3. Open the Crystal Data window for the first record and click the Comment tab. You see the information on the cluster composition:
that means that atoms Zn1 and Zr1 in the underlying nets correspond to 13-atom and 17-atom clusters, respectively. Then go to the Adjacency Matrix tab and be sure that the cluster centered by Zn1 is in contact with 12 other clusters (6Zn1+6Zr1) while the other cluster is linked to 16 clusters (12Zn1+4Zr1).

Open the Crystal Data window for the second record to be sure that one of two clusters is “empty” (ZA1), moreover, some intercluster contacts in the adjacency matrix are marked as “H bonds”. Pay attention that in the underlying net each “valence” contact corresponds to a pair of “tangent” clusters having at least one common atom, while “H bond” contact designates “bounded” clusters that are connected only by bonds between their surface atoms.

4. Close the Crystal Data window and run ADS for the first record in the Classification mode. You will find that the underlying net has the topology of the cubic Laves phase MgCu2 (mgc-x). The topology of other two underlying nets can be analyzed in two ways: ignoring or not ignoring links between “bounded” clusters. For this purpose you can set the Bond type option for H bonds to None or to At., respectively.

Check that for the second underlying net the two possible topologies are diamond (dia) and b.c.c. (bcu-x), respectively, while for the last underlying net the topologies are unknown to ToposPro.
5. The beginning of the Samson.txt file looks like:

6. Turn to the database Samson and run IsoCryst for the ZrZn22 record. Draw the picture and select all Zr1 and Zn3 atoms.

Leave the centers of clusters only and press one time the Growth button to be sure that the whole structure can be constructed with one-shell clusters centered by Zr1 and Zn3 atoms. Select Zr1 atoms only and show their coordination polyhedra (


7. Pay attention that the number of the NaCd2 nanocluster representations is much larger (41), but there is only one with two clusters (the simplest one). Select this representation only and construct its underlying net, be sure that the topology of the net is also mgc-x if both tangent and bounded cluster contacts are taken into account.

Open Samson.txt and be sure that both clusters are two-shell: Na1: 1@16@44 (61-atom cluster) and Cd6: 1@12@50 (63-atom cluster).

Draw both clusters in IsoCryst and look how the atoms of the second shell are allocated relative to the atoms of the inner polyhedron:


8. Select both records in the Samson database and open the IsoTest program. Click the option Common/Common Flags and indicate the Save Topology and Cont.Calc. mode. Then specify the following IsoTest options in Topology tab to compute coordination sequences for all atoms up to the 3rd coordination shell and store the information into the Samson.its file:


Run IsoTest and close its window after finishing the calculation. Then right-click in the database window and specify Show ITS to show information about coordination sequences in the Crystal Data window.

Open the Crystal Data window and go to the Structure Topology tab to see the coordination sequences.

Close the Crystal Data window and open the Distribution window (Database/Distribution). In the Distribution Type option set Sublatice Topology and press the Create Distribution button to construct the distribution of the coordination sequences.

Order the list by Property.

You see, for instance, that the clusters 1@12@50 allow four possible compositions for the third shell with 109, 113, 114 or 120 atoms, moreover, all of them occur in NaCd2 (you may press the Select button to select the corresponding records in the database).
9. Using the Crystal Data window, find that the cluster 1@12@50@113 is realized for Cd10 and Cd12 atoms of NaCd2. Open IsoCryst and construct three-shell cluster for Cd10 by two successive clicks of the Growth button. Select all atoms in the cluster and use Save Selection command (Graph files (*.gph) type of files) to store the cluster topology in the 1_12_50_113.gph file.

10. Open IsoCryst for the NaCd2 record, draw picture and use the Load Tint File command (Graph files (*.gph) type of files) to choose the 1_12_50_113.gph file to find that there is only one independent cluster of this type.

Press the Apply button to show visible fragment of the cluster, press the IsoCryst Growth button several times until the Fragment item in the list becomes bold (all atoms are visible).

Press the Apply button again and center the image (Space key) to see the cluster required.

Exercise: perform the same analysis for the crystal structure of β-Mg2Al3 (Mg2Al3.cif). What are the relations between this structure and the structures of ZrZn22 and NaCd2? Note: also use MinOm=0.00 value in Nanoclustering window.
(The nanocluster model can be presented as a combination of 1@16@44 and 1@12@50 nanoclusters with the Mg9 and Al14 centers, respectively. The topology of the net is mgc-x).

 nanocluster (1@16@44) |
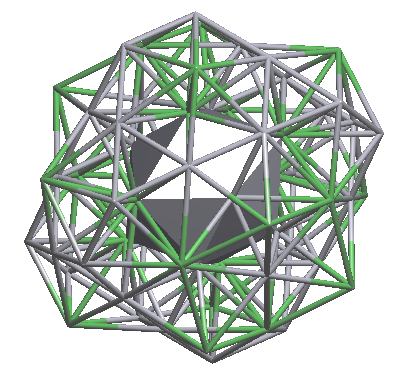 nanocluster (1@12@50) |